Hematopoiesis & Malignancies
The blood system is mainly maintained by continued hematopoiesis derived from hematopoietic stem cells (HSCs) during our life time. It is now a well studied model to investigate the regulatory network underlying lineage choice (cell fate decision), stem cell heterogeneity and clonal evolution in aging or blood malignancies. Single-cell techniques offer unprecedented opportunities to enable decoding these basic questions.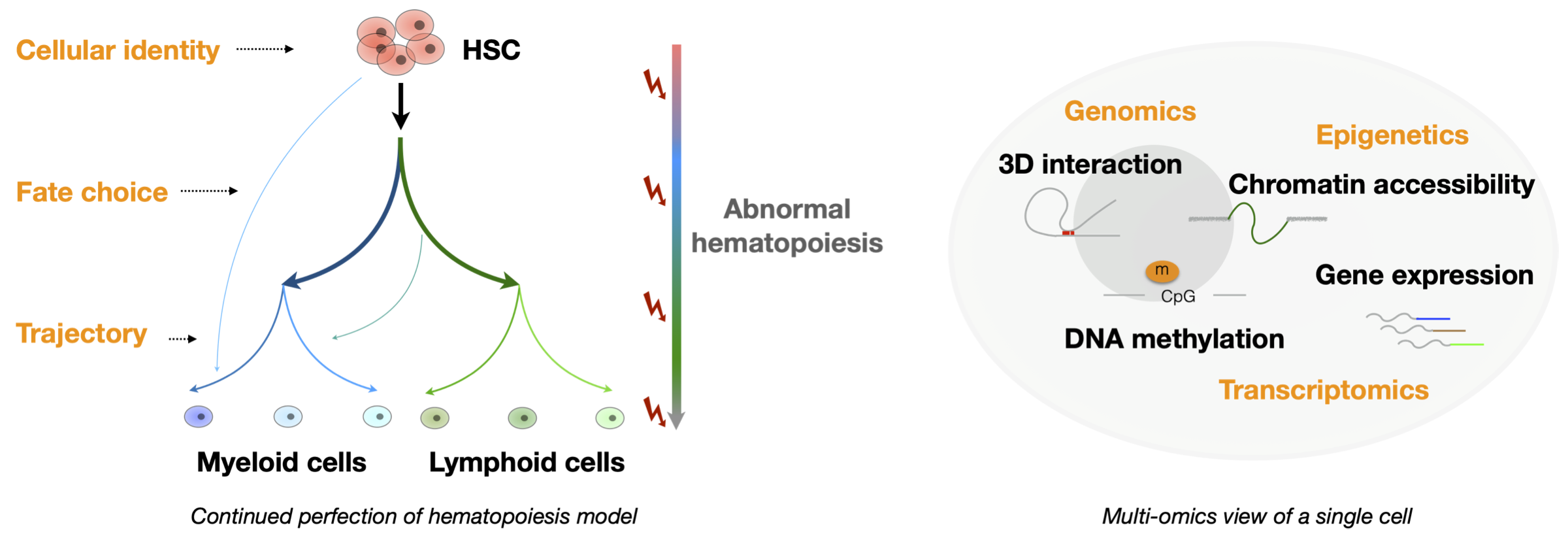 Focusing on hematopoiesis and malignancies, our lab aims to explore the systematic regulation of genetics, transcriptomics and epigenetics by combining single-cell multiomics techniques and bioinformatics. We anticipate that these studies will improve our understanding of the basic events during biological development, disease pathogenesis and resistance to therapies. Our final goal is to provide better strategies for the diagnosis, prediction and treatment of clinical diseases.
Focusing on hematopoiesis and malignancies, our lab aims to explore the systematic regulation of genetics, transcriptomics and epigenetics by combining single-cell multiomics techniques and bioinformatics. We anticipate that these studies will improve our understanding of the basic events during biological development, disease pathogenesis and resistance to therapies. Our final goal is to provide better strategies for the diagnosis, prediction and treatment of clinical diseases.
Hematopoiesis
[#Homeostasis and malignancies]
It is still elusive about the heterogeneous composition of HSPCs, and more importantly their relations to functional diversities, which are more announced in aging and blood diseases, especially malignancies. A major focus in the lab is the heterogeneity and functional relevance in HSPCs.
Single-cell Omics
[#Single cell resolution]
Heterogeneities are represented by multiple dimensions of characteristics and could not be completely explained at the bulk level. To better resolve the molecular underpinnings, we use single-cell techniques to study the heterogeneity of the genome, epigenome and transcriptome and other dimensions.
Bioinformatics
[#NGS multiomics]
Another interest in the lab is to pave the way of biological data mining by developing bioinformatics tools for data analysis and visualization, especially for single-cell sequencing data. See Tools page for the details of our previously developed tools.