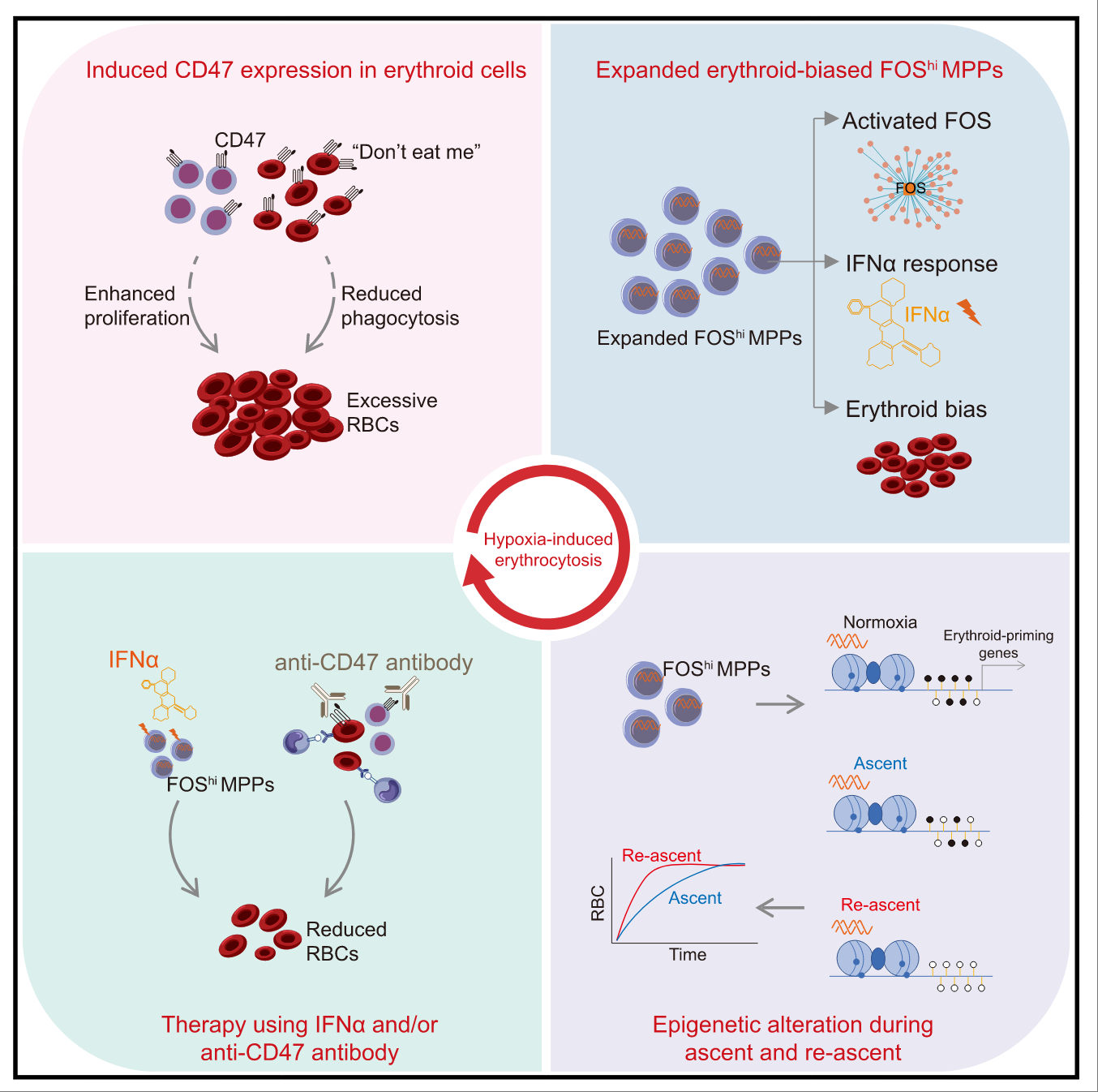
FOS-high erythroid progenitors contribute to adaptation to hypoxia
We characterized the cellular and molecular features of hematopoietic lineage cells under longterm hypoxia exposure using scRNA-seq. We identified a FOShi MPP subset with erythroid-lineage priming, as well as increased CD47 expression in erythrocytes, which enhances survival by evading phagocytosis. These two factors contribute to the hypoxia adaptation. Cell Stem Cell. 2025.
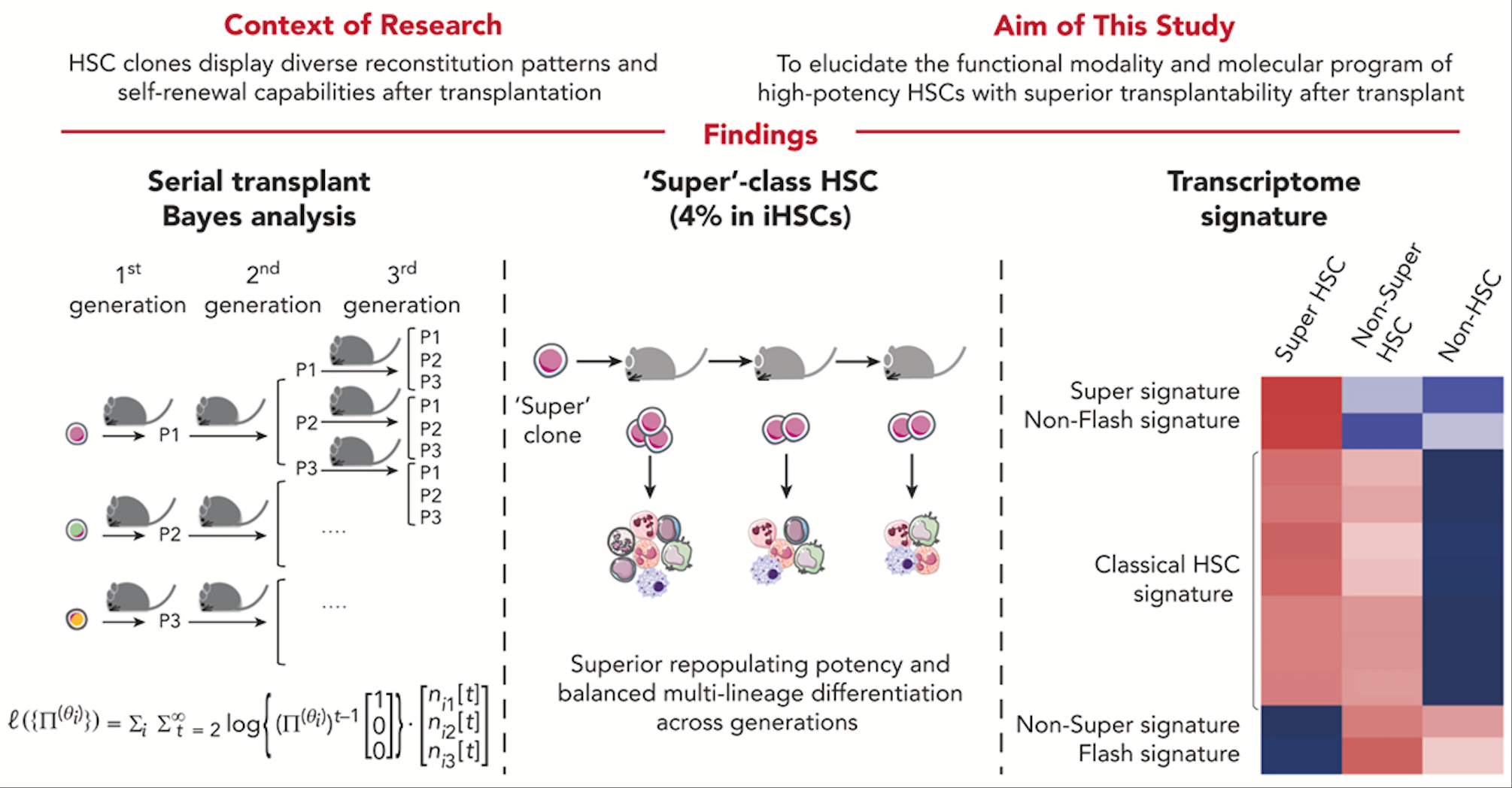
Identification of 'Super' HSCs in mice
'Super'-class HSCs were identified through functional analysis and Bayesian modeling. Molecular features of 'Super'-class HSCs were revealed by single-cell transcriptional analyses of their reconstituted HSCs. Blood. 2025.
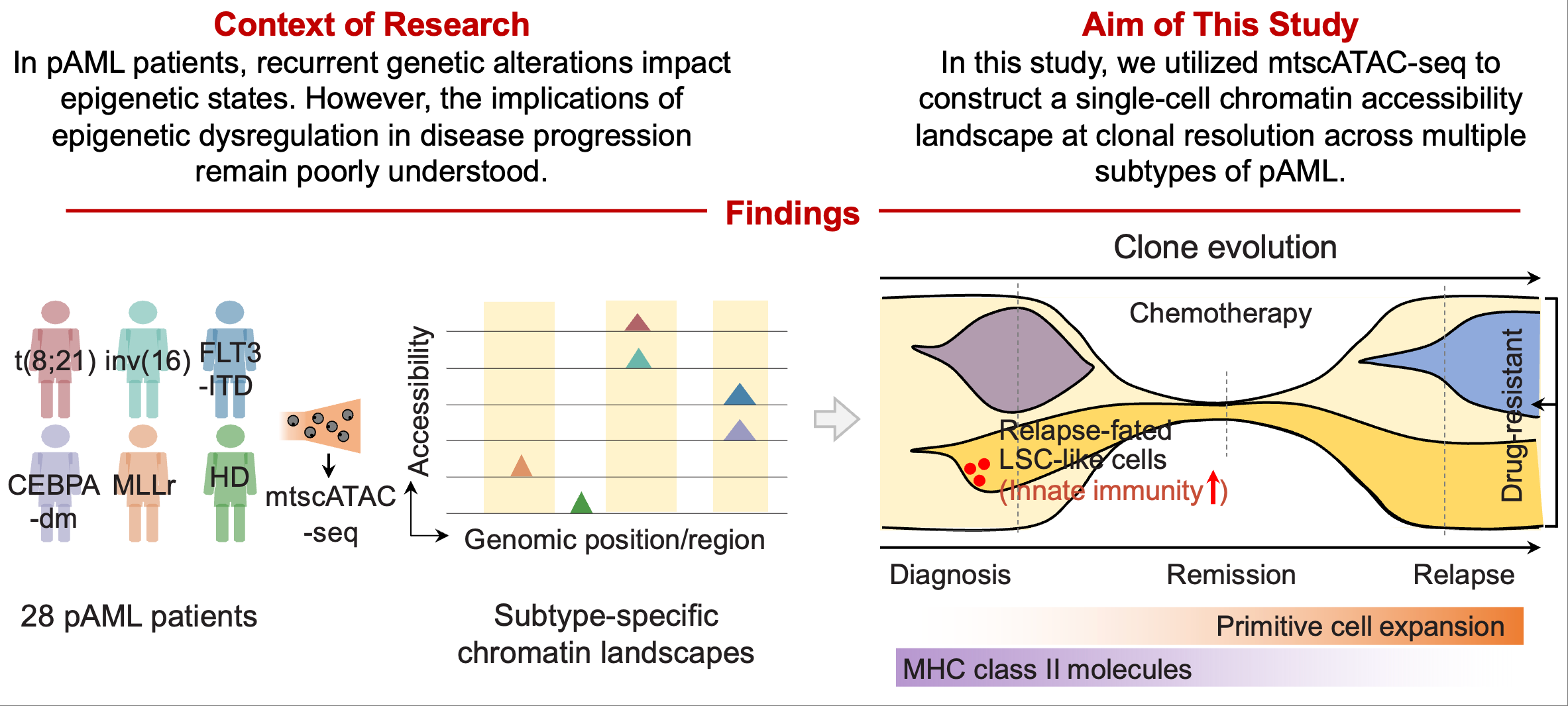
Epigenetic and clonal evolution in pAML
In pAML patients, a single-cell and clonal resolution map of chromatin accessibility reveals aberrant epigenetic kinetics from diagnosis to relapse. The reduction of MHC class II signaling in progenitor cells during relapse suggests a potential immune evasion mechanism facilitating clonal expansion. Blood. 2024.
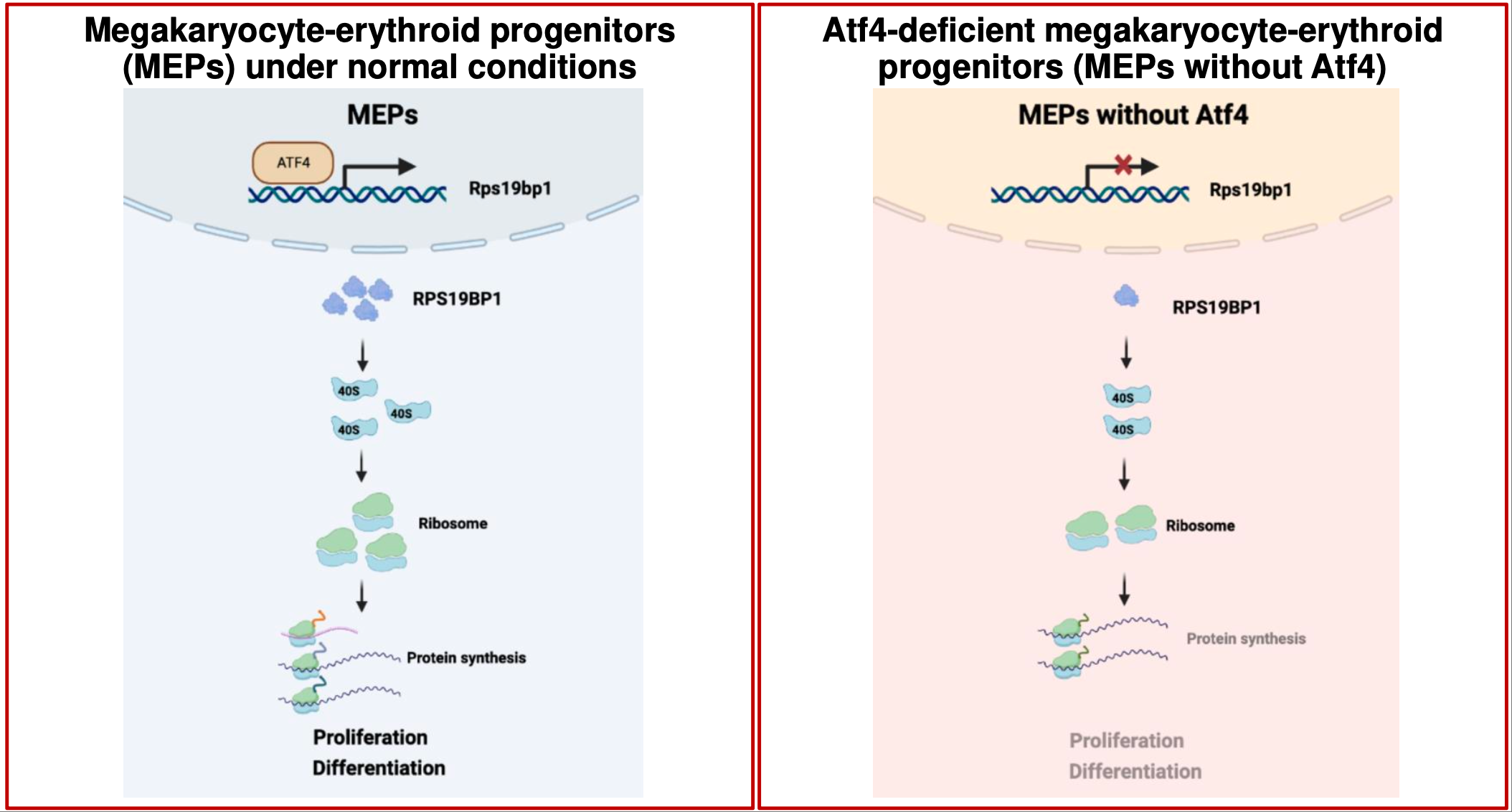
Activating ATF4 and regulation of erythropoiesis
In hematopoietic cells, ATF4 directly regulates the transcription of Rps19bp1, which in turn coordinates ribosome biogenesis promoting erythropoiesis. Hematopoietic cells that are deficient in Atf4 exhibit impaired self renewal and defective erythroid differentiation. Blood. 2024.
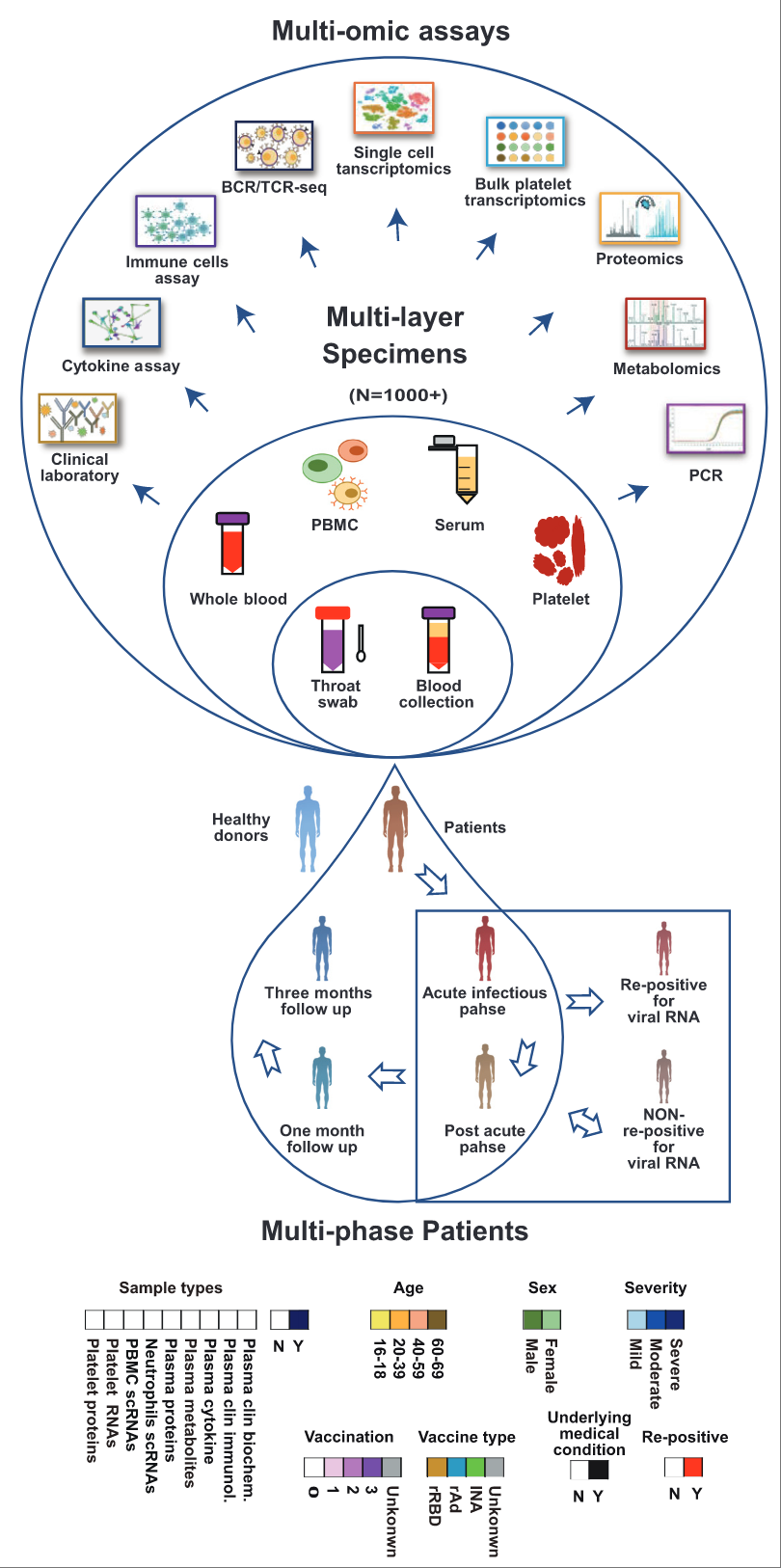
Multi-omics blood atlas during Omicron breakthrough infection
The blood parameters altered by SARSCoV-2 Omicron infection have not been sufficiently resolved. We profile more than 1,000 human blood or plasma samples to generate a multi-omics atlas, revealing unique features of the systemic host response and facilitating the development of screening and therapeutic strategies for Omicron breakthrough infection. Immunity. 2023.
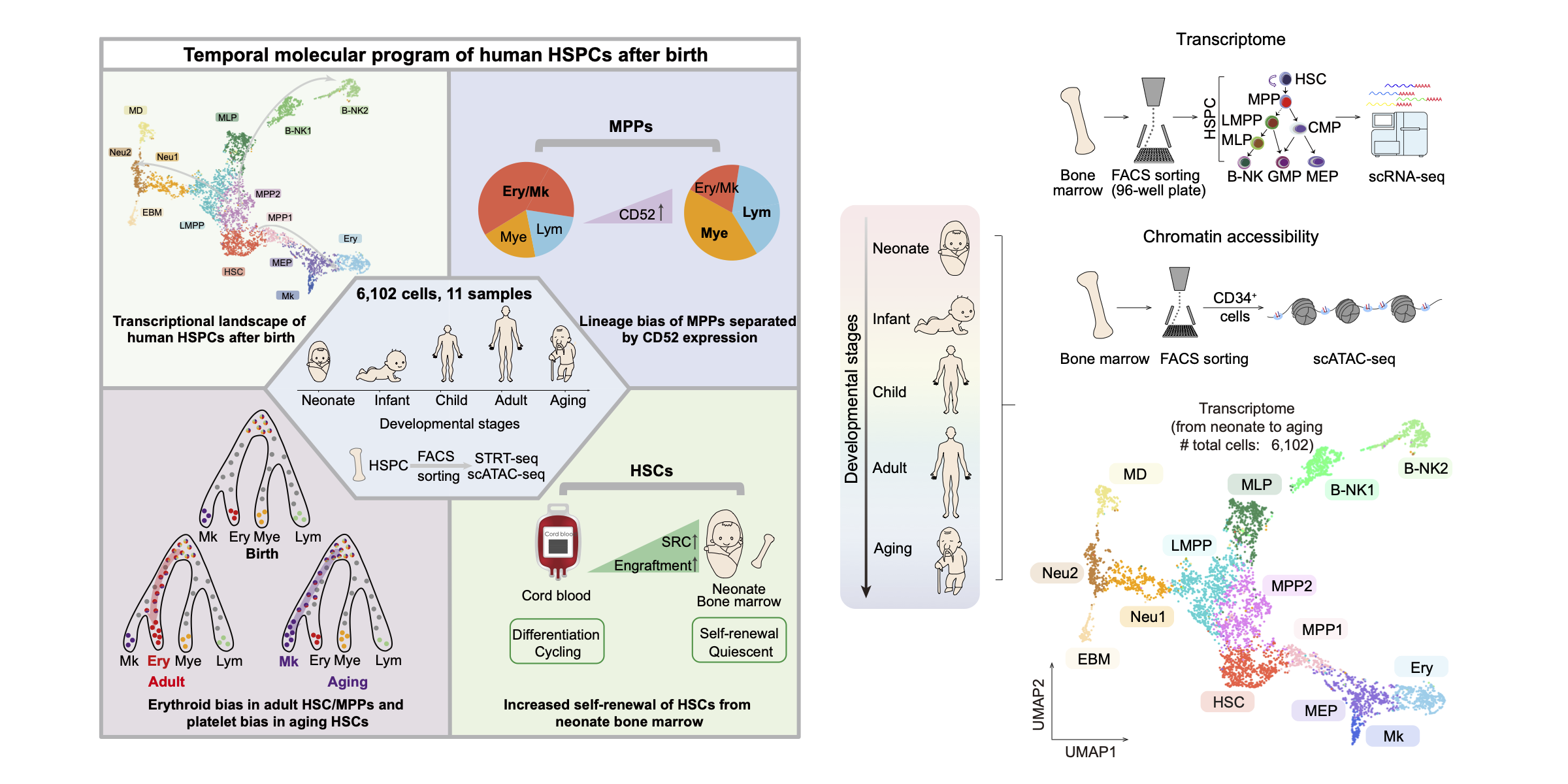
Molecular program of human HSPCs after birth
We provided an integral transcriptomic analysis of HSPCs, defined their dynamics in postnatal steady-state hematopoiesis and refined the heterogeneity of MPPs by CD52 expression. Neonate bone marrow-derived HSCs exhibited higher reconstruction capability than cord blood HSCs. This resource will help understand hematopoiesis in development and disease. Developmental Cell. 2022.
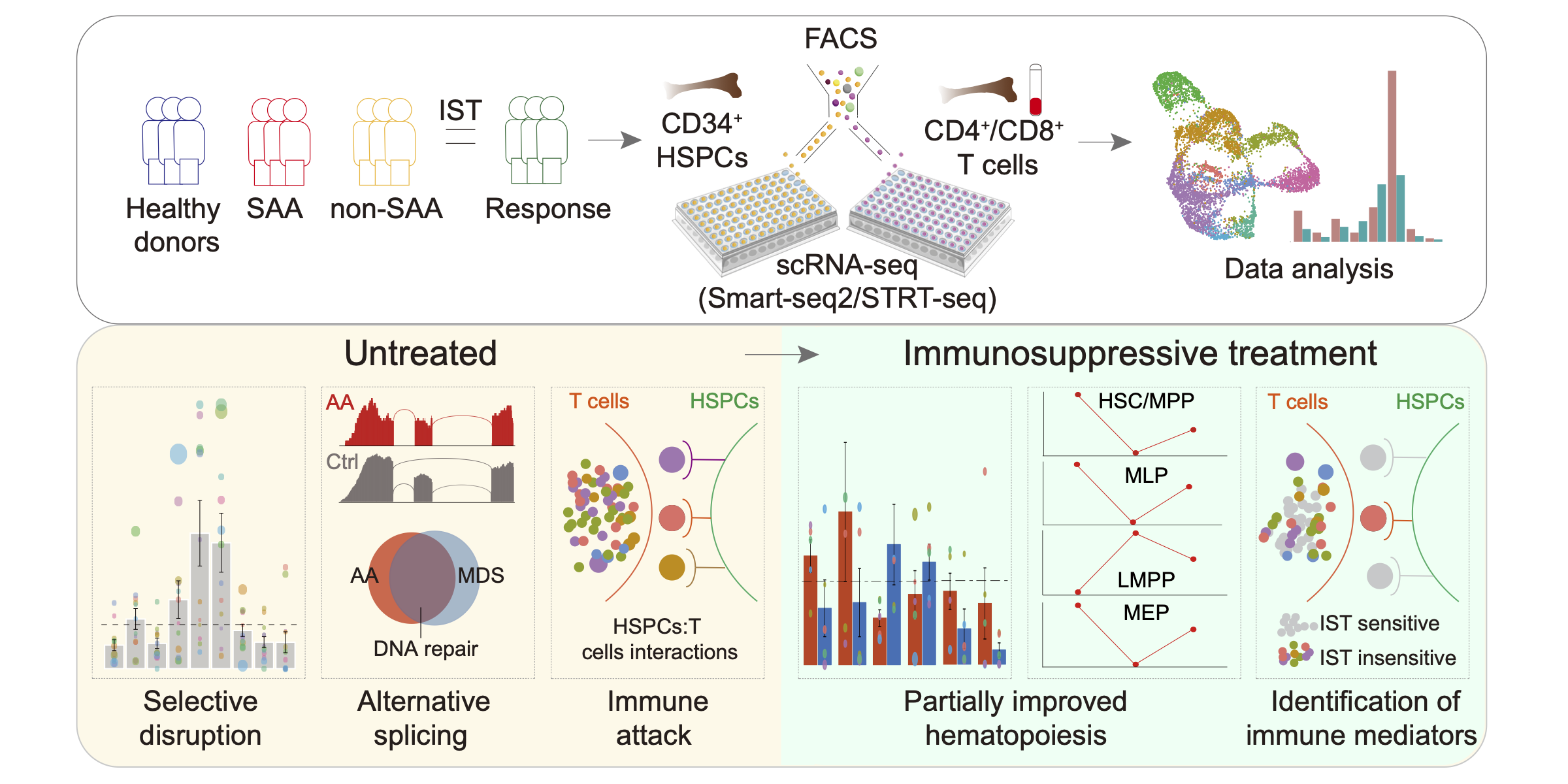
HSPCs and T cells in Aplastic anemia
These data collectively constitute the transcriptomic landscape of disrupted hematopoiesis in AA at single-cell resolution, providing new insights into the molecular interactions of engaged T cells with residual HSPCs and render novel therapeutic opportunities for AA. Blood. 2021.
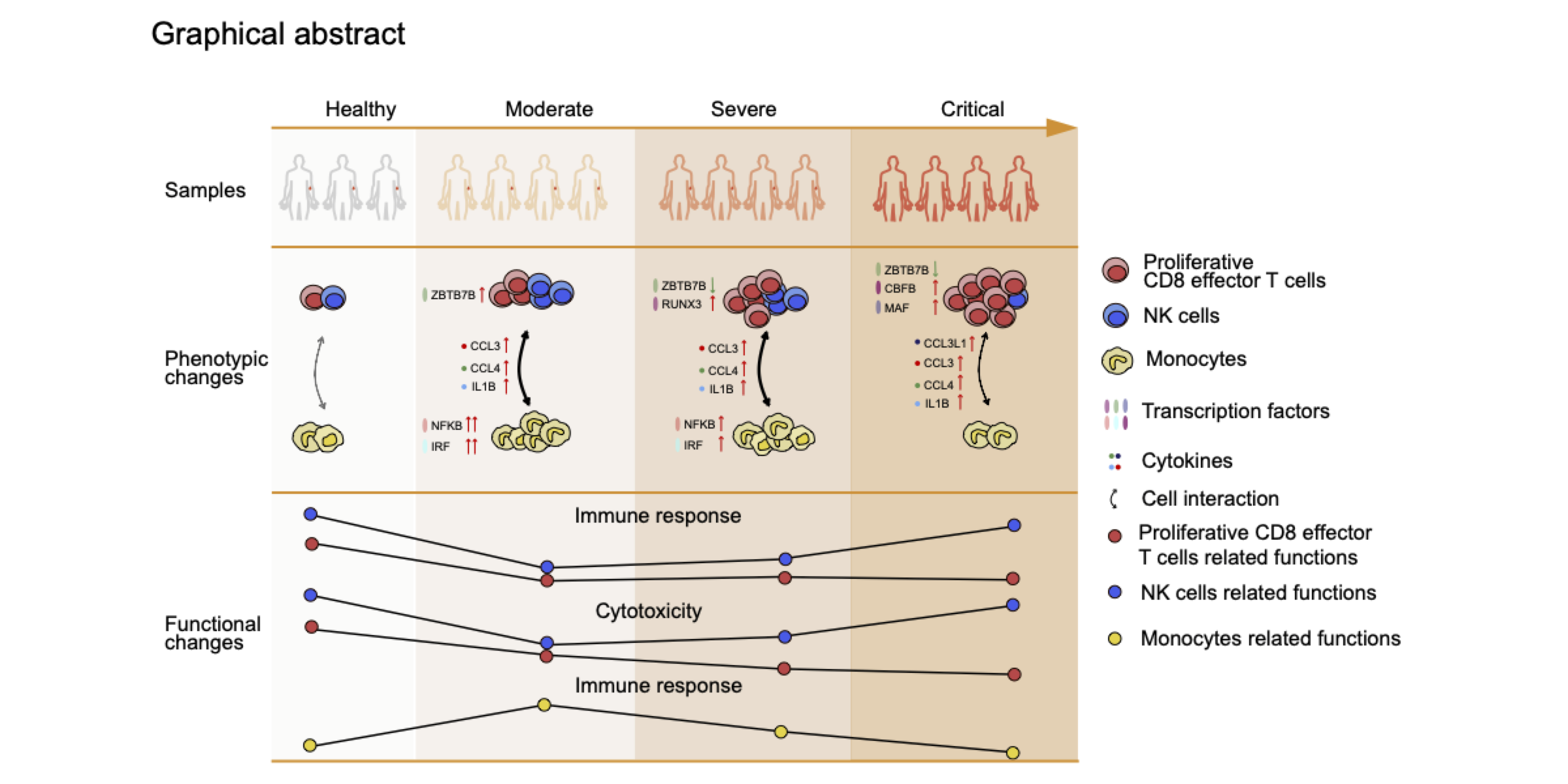
COVID-19 severity represented by blood cells
Our work uncovers the cellular and molecular players underlying the disordered and heterogeneous immune responses associated with COVID-19 severity, which could provide valuable insights for the treatment of critical COVID-19 patients. Sci China Life Sci. 2021.
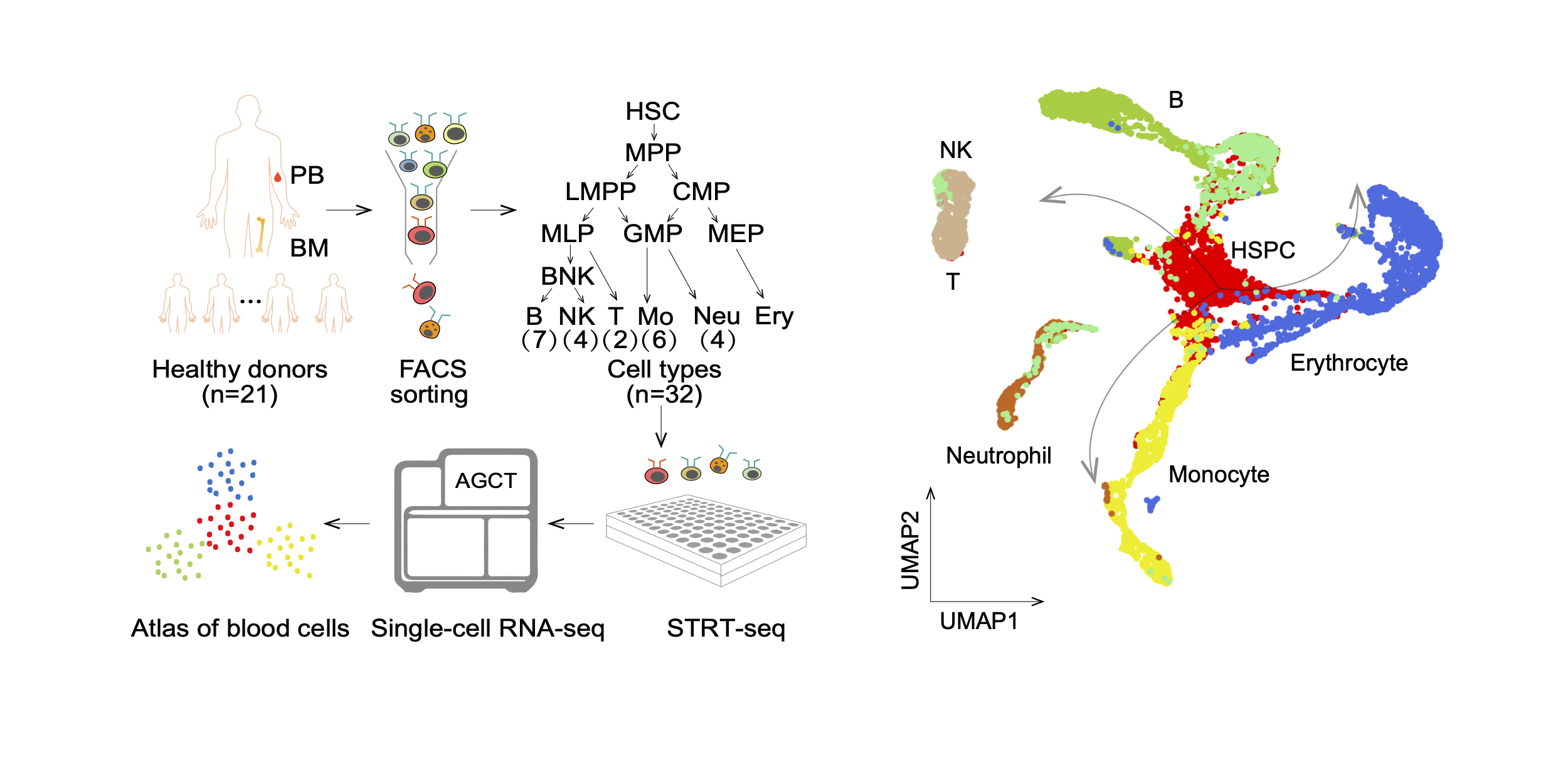
Atlas of Human Blood Cells
By in-depth sequencing and analysis of 32 immunophenotype-enriched blood cells at single-cell resolution, we built the transcriptional atlas of human blood cells. National Science Review. 2020.
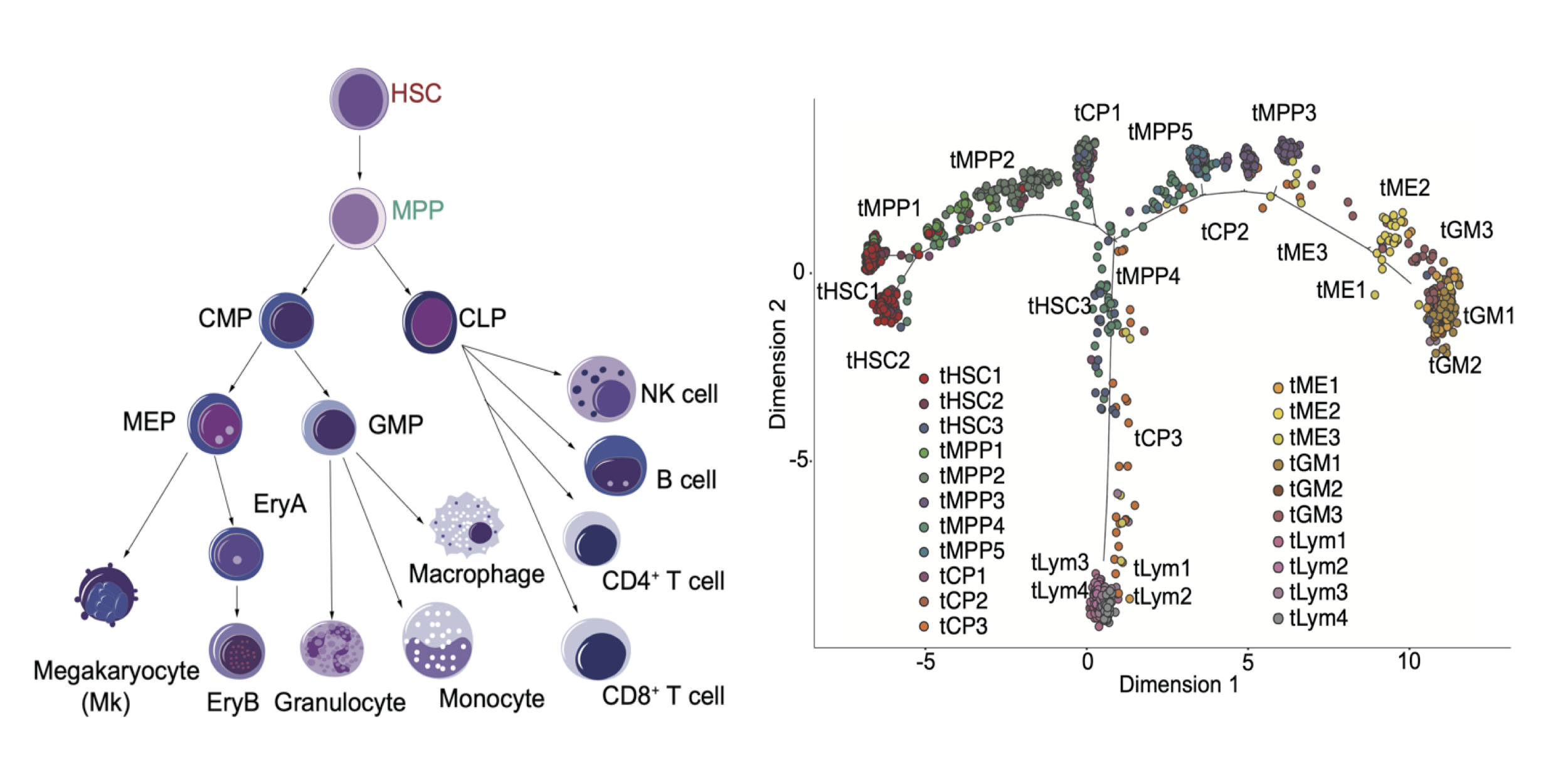
HSC Transplantation
We used single-cell RNA-seq to construct a comprehensive transcriptome reference in mouse homeostasis and guide the tracing of HSC transplantation dynamics in early one week window. Nature Cell Biology. 2020.
